Larus armenicus
Larus armenicus
(last update:
Amir Ben Dov (Israel)
Mars Muusse (Netherlands)
armenicus 1cy July
armenicus 1cy August
armenicus 1cy Sept
armenicus 1cy Oct
armenicus 1cy Nov
armenicus 1cy Dec
armenicus 2cy Jan
armenicus 2cy Febr
armenicus 2cy March
armenicus 2cy April
armenicus 2cy May
armenicus 2cy June
armenicus 2cy July
armenicus 2cy August
armenicus 2cy Sept
armenicus 2cy Oct
armenicus 2cy Nov
armenicus 2cy Dec
armenicus 3cy Jan
armenicus 3cy Febr
armenicus 3cy March
armenicus 3cy April
armenicus 3cy May
armenicus 3cy June
armenicus 3cy July
armenicus 3cy August
armenicus 3cy Sept
armenicus 3cy Oct
armenicus 3cy Nov
armenicus 3cy Dec
armenicus sub-ad Jan
armenicus sub-ad Febr
armenicus sub-ad March
armenicus sub-ad April
armenicus sub-ad May
armenicus sub-ad June
armenicus sub-ad July
armenicus sub-ad August
armenicus sub-ad Sept
armenicus sub-ad Oct
armenicus sub-ad Nov
armenicus sub-ad Dec
armenicus adult Jan
armenicus adult Febr
armenicus adult March
armenicus adult April
armenicus adult May
armenicus adult June
armenicus adult July
armenicus adult August
armenicus adult Sept
armenicus adult Oct
armenicus adult Nov
armenicus adult Dec
Dorit Liebers & Andreas J. Helbig
published: Limicola 13-6, 1999 (in German, with English summary)
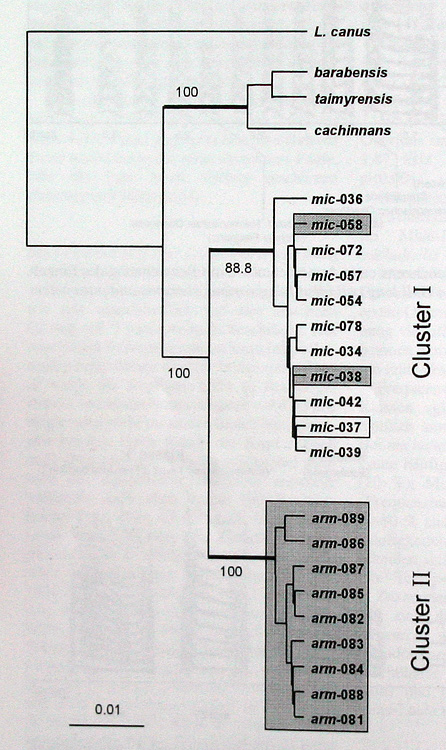
Fig 24: Reconstruction of geneology tree for armenicus and michahellis.
results on mitochondrial DNA in armenicus
At the three sites in Turkey, 97 gulls (10 adults at Tuz Golu plus 87 chicks) were sequenced and 12 different haplotypes (sequences that were different from ach other in at least one nucleotide) were found. These 12 haplotypes are the framed numbered haplotypes in Fig. 24. Three of these 12 haplotypes were identical to haplotypes found in michahellis from the E. Mediterranean, but the other 9 haplotypes were only found in the interior colonies of Turkey.
The reconstructed genealogy (the evolutionary ancestry tree) of the michahellis/armenicus-haplotypes shows two distinct clusters. One cluster (Cluster II) consists of 9 haplotypes that were only found in armenicus at Tuz and Van Golu. The other cluster (Cluster I) consists of 11 haplotypes which all can be found in michahellis from the E Mediterranean but were also found at Tuz Golu (2 haplotypes) and at Beysehir Golu (one haplotype).
The frequency of occurrence of these two cluster of haplotype-groups over the colonies is represented in Fig. 25 (Cluster I is white; Cluster II is black). It is immediately obvious that there were no armenicus haplotypes found in michahellis breeding along the coast of the Mediterranean or the Black Sea (but more study east of Crete and larger sample sizes are needed). The reverse is obvious as well: michahellis haplotypes do occur in colonies in interior Turkey, even in the phenotypically ‘pure’ armenicus colony of Tuz Golu (14% of the examined birds from this population show michahellis-type haplotypes). In the phenotypically mixed colony of Beysehir, even 62% of the birds have michahellis-type haplotypes. At Van Golu, where 30 birds (chicks from different nests) were examined, no such haplotypes were found (but this does not completely exclude the possibility of occurrence at this site; a larger sample size should reveal this).
Phylogenetically, Armenian Gull was found to be most closely related to michahellis to the exclusion of taymirensis, barabensis and cachinnans (Fig. 24). Michahellis and armenicus are represented by two distinct clusters in the mitochondrial haplotype phylogeny, but introgression (unidirectional geneflow) was evident with two michahellis haplotypes (shaded in cluster I, Fig. 24) having invaded the armenicus population at Tuz Golu. These and a third michahellis haplotype were found in 62% of the birds in Beysehir, 14% at Tuz and 0% at Van Golu (Fig. 25). This indicates that hybridization does occur, hybrids are fertile and backcross at least with armenicus in central Anatolia. Whether geneflow in the other direction (into the michahellis population of the E Mediterranean) also occurs requires further study. Notwithstanding the ongoing introgression, the phenotypically “pure” armenicus population at Tuz and Van Golu was genetically significantly differentiated from michahellis (AMOVA: ФST = 0.7857; p < 0.0001). The genetic divergence of the two haplotype clusters (average 2,05% at 420 nucleotides of control domain I, Fig. 26) indicates a relatively ancient separation and long independent evolution between michahellis and armenicus, but this has so far not resulted in full reproductive isolation.